-Search query
-Search result
Showing 1 - 50 of 136 items for (author: ruan & j)

EMDB-17740: 
Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5
Method: single particle / : Kobayashi W, Sappler A, Bollschweiler D, Kummecke M, Basquin J, Arslantas E, Ruangroengkulrith S, Hornberger R, Duderstadt K, Tachibana K

EMDB-17741: 
Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5
Method: single particle / : Kobayashi W, Sappler A, Bollschweiler D, Kummecke M, Basquin J, Arslantas E, Ruangroengkulrith S, Hornberger R, Duderstadt K, Tachibana K

PDB-8pki: 
Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5
Method: single particle / : Kobayashi W, Sappler A, Bollschweiler D, Kummecke M, Basquin J, Arslantas E, Ruangroengkulrith S, Hornberger R, Duderstadt K, Tachibana K

PDB-8pkj: 
Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5
Method: single particle / : Kobayashi W, Sappler A, Bollschweiler D, Kummecke M, Basquin J, Arslantas E, Ruangroengkulrith S, Hornberger R, Duderstadt K, Tachibana K

EMDB-29794: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29795: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29796: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29797: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g72: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g73: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g74: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g75: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-35782: 
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Method: single particle / : Ruan MX, Ding W

EMDB-35783: 
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Method: single particle / : Ruan MX, Ding W
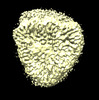
EMDB-35784: 
Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value
Method: single particle / : Ruan MX, Ding W

EMDB-35785: 
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Method: single particle / : Ruan MX, Ding W

EMDB-35786: 
Cryo-EM structure of protonated LHCII nanodisc at low pH value
Method: single particle / : Ruan MX, Ding W
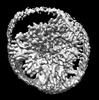
EMDB-35787: 
Cryo-EM structure of unprotonated LHCII nanodisc at low pH value
Method: single particle / : Ruan MX, Ding W

PDB-8iwx: 
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Method: single particle / : Ruan MX, Ding W

PDB-8iwy: 
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Method: single particle / : Ruan MX, Ding W

PDB-8iwz: 
Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value
Method: single particle / : Ruan MX, Ding W

PDB-8ix0: 
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Method: single particle / : Ruan MX, Ding W

PDB-8ix1: 
Cryo-EM structure of protonated LHCII nanodisc at low pH value
Method: single particle / : Ruan MX, Ding W

PDB-8ix2: 
Cryo-EM structure of unprotonated LHCII nanodisc at low pH value
Method: single particle / : Ruan MX, Ding W

EMDB-33845: 
Cryo-EM map of Rpd3S complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33846: 
Cryo-EM map of Rpd3S:head-bridge-right arm
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33847: 
Cryo-EM map of Rpd3S:bridge-left arm
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33848: 
Cryo-EM map of Eaf3 CHD bound to H3K36me3 nucleosome
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33849: 
Cryo-EM map of Rpd3S in loose-state Rpd3S-NCP complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33850: 
Cryo-EM map of Rpd3S in close-state Rpd3S-NCP complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33851: 
Cryo-EM map of Rpd3S complex bound to H3K36me3 nucleosome in close state
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33852: 
Cryo-EM msp of Rpd3S complex bound to H3K36me3 nucleosome in loose state
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

PDB-7yi0: 
Cryo-EM structure of Rpd3S complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

PDB-7yi1: 
Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

PDB-7yi2: 
Cryo-EM structure of Rpd3S in loose-state Rpd3S-NCP complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

PDB-7yi3: 
Cryo-EM structure of Rpd3S in close-state Rpd3S-NCP complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

PDB-7yi4: 
Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

PDB-7yi5: 
Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-28825: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

EMDB-28827: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

PDB-8f2r: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

PDB-8f2u: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

EMDB-28087: 
CryoEM structure of GSDMB in complex with shigella IpaH7.8
Method: single particle / : Wang C, Ruan J

EMDB-28583: 
CryoEM structure of GSDMB pore without transmembrane beta-barrel
Method: single particle / : Wang C, Ruan J

EMDB-28584: 
CryoEM structure of the GSDMB pore
Method: single particle / : Wang C, Ruan J

PDB-8efp: 
CryoEM structure of GSDMB in complex with shigella IpaH7.8
Method: single particle / : Wang C, Ruan J

PDB-8et1: 
CryoEM structure of GSDMB pore without transmembrane beta-barrel
Method: single particle / : Wang C, Ruan J

EMDB-28535: 
Human PAC in nanodisc at pH 4.0 with PI(4,5)P2 diC8
Method: single particle / : Ruan Z, Lu W

EMDB-28964: 
Human PAC in nanodisc at pH 4.0 with PI(4,5)P2 diC8
Method: single particle / : Ruan Z, Lu W
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
